During a lecture on the basics of medical imaging some time ago I, together with a classmate, wrote a simple script for ImageJ (An image editor and processor implemented in Java) that achieves a HU-threshold based segmentation of a chosen region.
While the ImageJ’s scripting functionality is quite versatile, the example showed that it suffers from a major drawback, which is speed. The main reason for this is that in order to leave the original DICOM-image unchanged, a second image is created as buffer to label the region and a constant switch between the two images is required which actually slows down the whole procedure quite a bit. Still, this seemed the more reasonable option compared to painting into the actual DICOM-image and potentially ruining information. Using the buffer image allows restarting the segmentation by simply choosing a new seedpixel and comparing different results easily.
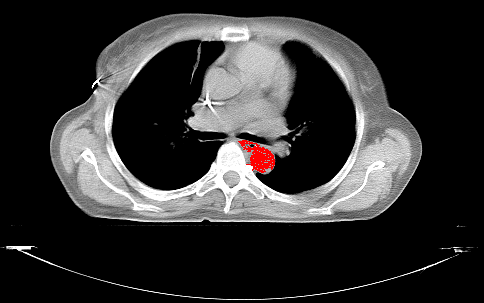
Before the basic functionality of the script is explained, here’s the result of a segmentation of the aorta in a single slice contained in a mamma-ct, which was used as testimage during the development of the script:
The script allows to set a threshold (which in DICOM-images represents the deviation from the seedpoint in HU that is still counted as belonging to the region) and upon activation uses the cursorlocation when clicking for determining the seedpoint, which is marked as region (red pixel) in the buffer image. It then starts checking the pixels in four directions for compliance with the regions thresholds, marks them accordingly, and recursively starts the checks in four directions (which is another reason for the script being slow, but implementing a more complex yet faster algorithm wasn’t really the target of writing this script).
The script can be found here.